A team of Hebrew University scientists and physicians has come up with a way to reduce wait-time for results and conserve precious laboratory supplies – without compromising test sensitivity or result validity.
As the Coronavirus began spreading, public health and medical professionals recognized the importance of quickly identifying, and isolating, people infected with SARS-CoV-2. This holds especially true for retirement homes, hospital staff, or workers at essential factories. There is a great need to quickly screen large groups of people, especially to rule out even the low probability of infection in individuals, thereby ensuring the safety of the work environment, essential to its functioning.
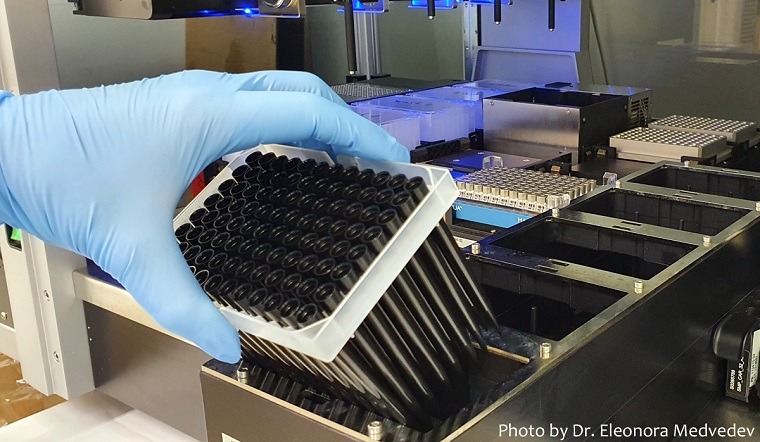
The standard polymerase chain reaction (PCR) test is conducted on genetic material extracted from patient swabs. Each patient’s sample has to be processed and tested individually: from inactivating the virus, extracting the genetic material, and conducting the positive/negative test. This requires substantial effort and cost of the materials for each sample. Furthermore, very early in the pandemic, world shortages of necessary materials became apparent, limiting the numbers of tests that could be conducted at once.
A team including Dr. Yotam Drier, Dr. Maayan Salton, and Prof. Yuval Dor of the Hebrew University’s Faculty of Medicine, together with Prof. Dana Wolf of the Hadassah Medical Center, rose to the challenge, by initiating effective pooled patient testing. The Hebrew University was among the first in the world to safely implement this approach, which is currently being adopted globally.
What is pooling? Say only three of 80 people to be tested actually carry the virus. This means that most tests will turn out negative. The researchers surmised that the 80 samples could be pooled into ten groups samples, each containing samples from 8 individuals. This “pool” can then be tested as if it were only one individual, thereby conducting 10 instead of 80 tests. If a particular pool comes back negative, it means that all eight people are uninfected and there is no need to test them individually. If, on the other hand, a pool comes back positive, that particular batch will be “unpacked” and retested, to identify the infected person. This results in substantial reduction in cost and time, particularly when the positive results are a low percentage of the tests, and allows much more cost-effective screening of large groups of people.
While the principle behind this approach sounds simple, it is in fact much more complex, both theoretically and practically. The researchers conducted robust mathematical and statistical modelling, providing the theoretical basis of why and how pooling would work, and determining the optimal pool sizes. Practically, pooling requires robots to be programmed to pool eight samples, and assurances must be in place to guarantee sample quality.
This method’s success has expanded the testing capacity of the joint Hebrew University-Hadassah virology lab, reaching several thousand tests a day, and consequently in Israel as a whole. The lab operates 24/7 under the leadership of Prof. Wolf, with Hebrew University students and volunteers running the tests and conducting the lab work.
"We are happy to be able to contribute to the national effort fighting COVID-19. The unique strengths of the Hebrew University, including expertise in molecular biology and computer sciences, the close cooperation with the Hadassah Medical Center, the availability of cutting-edge equipment, and the positive spirit of the university’s staff, students, and researchers – who volunteered by the hundreds – have made it possible for the Faculty of Medicine and Hadassah to turn our campus into a national leader of Coronavirus testing. We continue our R&D to further improve diagnoses and identify potential interventions."
Prof. Yuval Dor, Faculty of Medicine
Rising Infection Rates
Yet the efficiency of pooling goes down as the percentage of positive swabs goes up. As infection rates began rising in late June, it became increasingly likely that pools would test positively – requiring all eight swabs be tested individually. Dr. Moran Yassour, whose research bridges medicine and computer science, sees this first and foremost as a mathematical question.
"The optimal assignment of samples into pools will make or break this. We must mathematically predict a way to triage samples as they arrive at the lab, assigning each test its ideal testing scheme to minimize the number of tests while maximizing their sensitivity and specificity."
Dr. Moran Yassour
Dr. Yassour is evaluating the entire testing process, by working backwards. The joint Hebrew University-Hadassah virology lab currently has a collection of over 120,000 swabs; by analyzing data from multiple test features (e.g. the protocol used, machine type, specific extraction kit and other parameters) and correlating these with clinical variables (such as patient age, gender, and symptoms), Dr Yassour can develop algorithms to determine the exact combinations in which specific samples should be pooled with each other to ensure the highest efficiency. "This will allow us to simulate the effectiveness of different pooling techniques, and continuously recommend optimal pooling techniques for any given time,” Dr. Yassour says.
"The optimal assignment of samples into pools will make or break this. We must mathematically predict a way to triage samples as they arrive at the lab, assigning each test its ideal testing scheme to minimize the number of tests while maximizing their sensitivity and specificity."
Dr. Moran Yassour
Together with with Prof. Yuval Dor and Prof. Dana Wolf, Dr. Yassour would like to use this data to train a machine learning based model to predict the probability of a false negative.
For more details and a complete list of researchers involved in this project, see the publication in Clinical Microbiology and Infection.